Overview of Inter-Species Comparisons in Pathway ToolsPathway Tools offers an array of powerful functions. Some of these are available only in the desktop mode of operation (requires installing MetaCyc plus Pathway Tools locally), whereas others are available only in the Web mode of operation. Some capabilities are available in both modes. The following examples demonstrate some of these functions for inter-species comparisons. 1. Comparative Tables: A Diverse Array of Global Comparisons Description: Many tables containing comparative statistics of two different Pathway/Genome Databases can be generated through the Web mode of Pathway Tools. To access: use the Web command Tools -> Comparative Analysis. 2. Global Comparisons: Comparing Overall Metabolic Maps between Organisms Description: In the desktop version of PathwayTools, the Metabolic Overview mode can be used to highlight all reactions shared or not with any/all members of a group of organism databases specified by the user. This highlighting allows you to perform global comparative analyses, that is, comparing the metabolic overview of the current organism against those of one or more other organisms. Application: A typical use of such analyses is in the development of antimicrobial drugs. When an antimicrobial agent is designed to target a specific metabolic enzyme/reaction, this analysis can computationally predict the spectrum of action across the organisms for which databases are provided. |
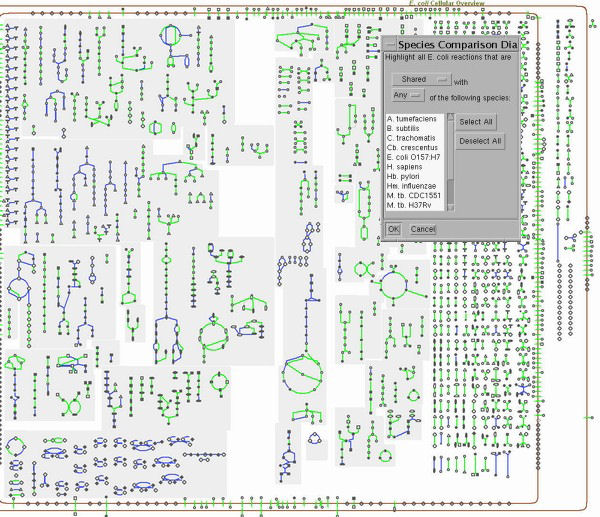
|
3. Cross-Species Comparisons: Comparing a Specific Pathway in Multiple Organisms Description: Both the desktop version and the Web version of PathwayTools support comparisons between the same pathway across two or more different organisms. To access: Click the "Cross-Species Comparison" button found at the top of all pathway pages. |
|
Application: Among many applications, this is useful following the computational pathway prediction of an organism, when one wishes to understand how a given pathway differs across two organisms. |

|
4. Cross-Species Comparisons: Comparing Genomic Maps Description: |
|
Application: This can be useful when attempting to understand differences in gene regulation between orthologous genes, where the difference might be attributable to inclusion in different operons or some other gene locus influence. |

|