MetaCyc Web Querying Examples |
|---|
Example 1: Using Text-Based Querying for an Object whose Name is UnknownSuppose you want to find information related to 6-phosphofructokinase but have forgotten its precise name. All you remember is that the enzyme is a kinase involving fructose. Step 1:Use the Quick Search box that appears at the top of every MetaCyc web page to search for all objects (proteins, reactions, genes, etc) that contain the words "kinase" and "fructose": |
|
Step 2: Hit "enter" or click the "Quick Search" button to see hits from MetaCyc proteins, Gene Ontology terms and reactions that match this query (below). From there, you can access extensive information about the proteins, as well as the reactions associated with phosphofructokinase, by clicking on individual items. 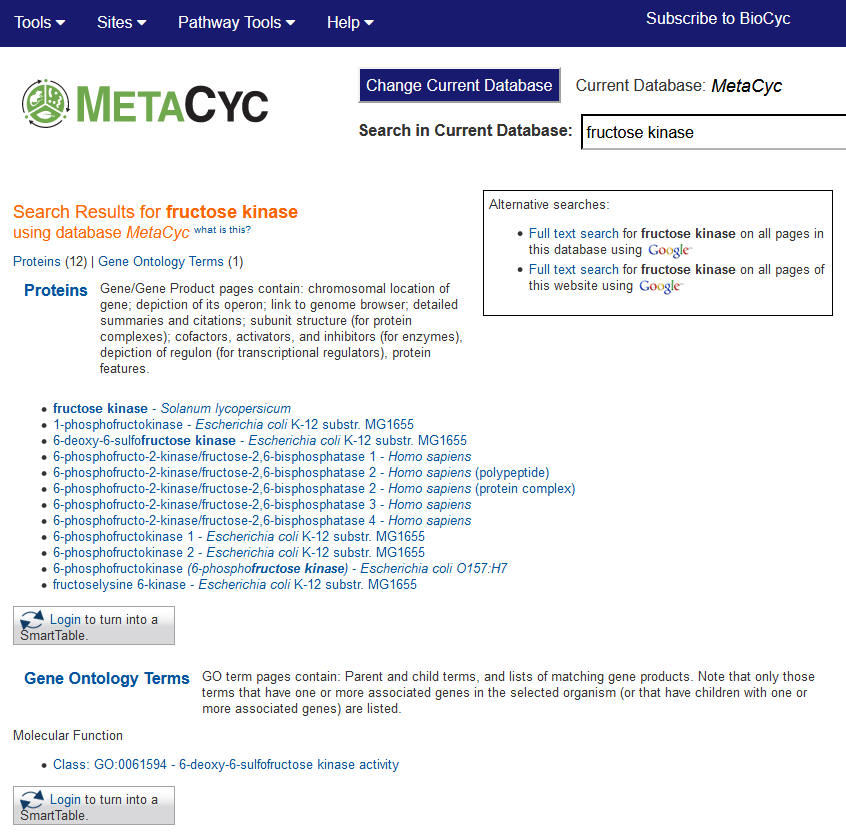 |
| [top] |
Example 2: Browsing MetaCyc using Ontologies for Pathways Related to Glycolysis[step 1 | step 2 | step 3 | step 4] Suppose you wanted to get an overall view of pathways involved in glycolysis. As part of MetaCyc's curation process [def], all of its pathways are classified according to a pathway ontology [def]. Step 1: Use Metabolism → Browse Pathway Ontologies command (from the Tools menu) to retrieve all high-level pathways (i.e., without retrieving every instance of a pathway subsumed under those headings): 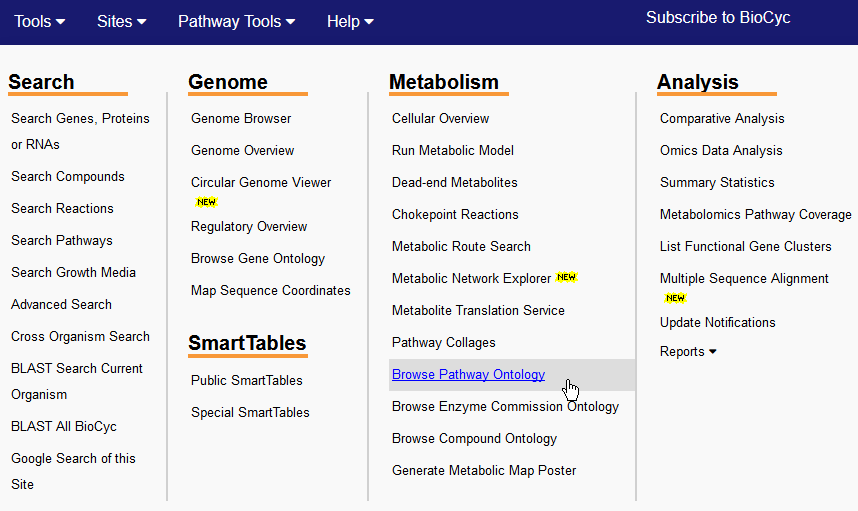
|
|
Step 2: In the resulting page, identify the appropriate root class (for glycolysis, this would be "Generation of precursor metabolites and energy", and expand it by clicking on the "plus" sign next to it. Once the list expands, click on Glycolysis: 
|
|
Step 3: Clicking on "Glycolysis" opens a window that lists the different pathways that are classified under glycolysis. One of the powerful features of MetaCyc is the ability to show multiple pathways covering the variability of metabolic pathways found in nature.  |
|
Step 4: Clicking the different links under the Glycolysis class will display the alternative glycolysis pathways found in different organisms. For example, clicking on Glycolysis V (Pyrococcus) will result in the following pathway. The summary section below the pathway diagram will provide information about the pathway, including the types of organisms in which this pathway variant is found. 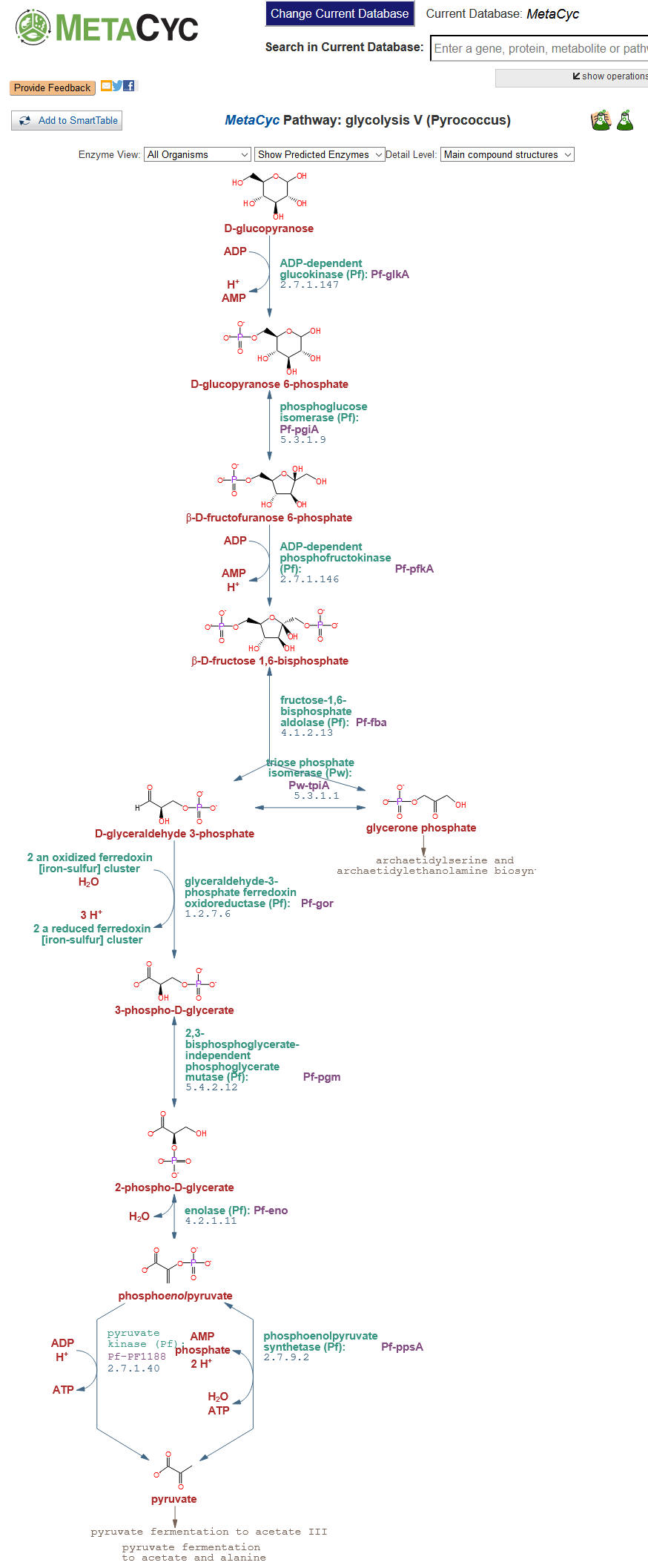 |
|
[top]
|
Example 3: Direct querying of MetaCyc using an IdentifierSuppose you know the name of a gene (e.g., fruK). Step 1: You can use either the Quick Search box at the top of the page, or (to avoid receiving results that are not genes) you can use the "Search Genes, Proteins orRNAs" command (from the Tools → Search menu). 
|
|
Step 2: When the MetaCyc "Gene/Protein/RNA Search" page appears, type fruK in the text field. As you type, the system will automatically suggest matches to the text entered so far; for example, after you typed "fru" the system may suggest genes such as FruB, fruI and fruK as possible candidates. After you type "fruK", only the fruK genes in MetaCyc appear as candidates. 
As this point you can either click on one of the suggestions before you submit your query (by clicking on the "Submit Query" button or hitting "Enter"), in which case you will be taken directly to that object's page, or you may ignore the suggestions, and submit the query as entered, in which case you will be taken to a page that lists all possible matches, as shown below. 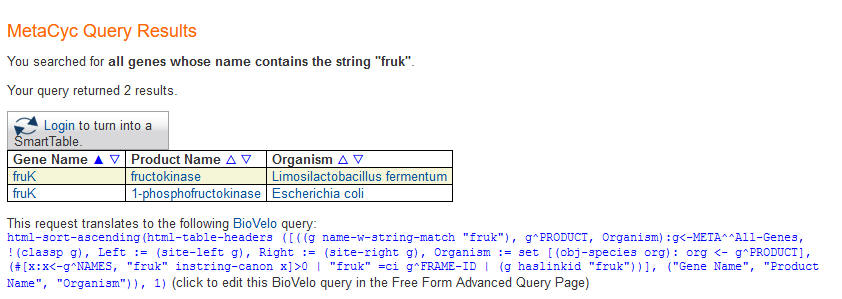
|
|
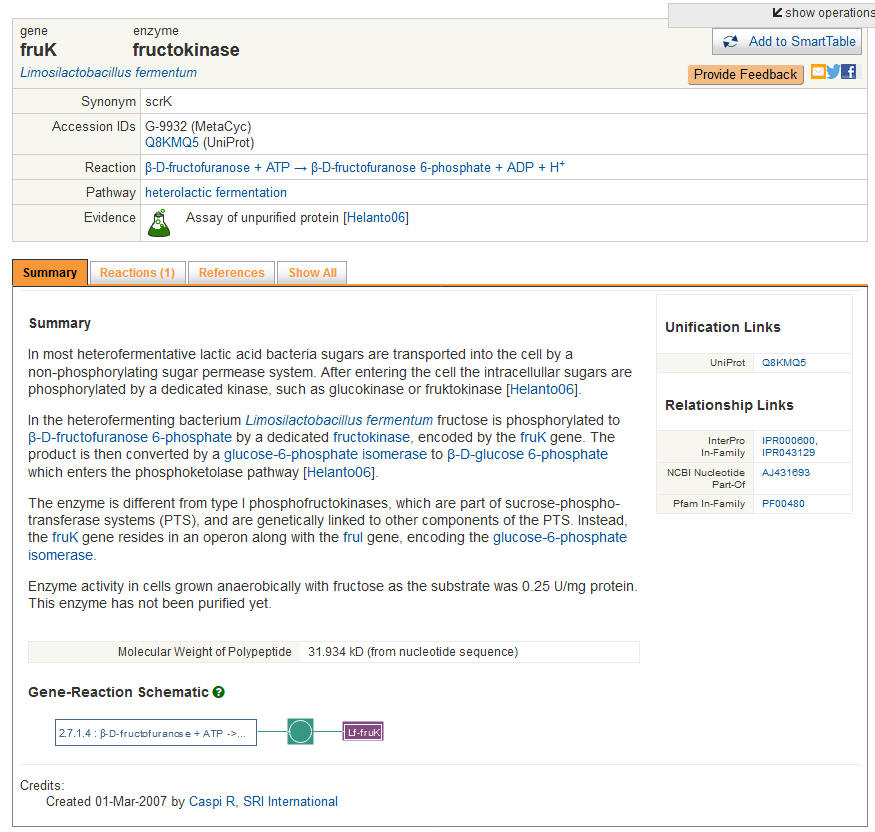
Outcome: Once you click on an individual gene, you will be taken to the MetaCyc's entry for that gene:
|
| [top] |
Desktop Application
Bring the power of BioCyc.org securely in-house, with the abilities to create and
edit PGDBs, perform metabolic modeling, and query/update using APIs.
Academic Licenses Commercial Licenses